Guided example
CXCL12 through ChemoPar-db
Follow this example to see how ChemoPar-db captures chemokine metadata, structures, and interaction fingerprints of CXCL12.
Protein page
Overview and numbering
- The header summarises alternate names, organism, identifiers, and direct links to UniProt and the Guide to Pharmacology.
- A scrollable sequence viewer aligns residues with the chemokine generic numbering, split by structural regions such as the N-loop, beta-sheets, and alpha-helix.
- A customisable 2D chemokine diagram adapts the GPCRdb snake plot to highlight CXCL12 topology and residue groupings.
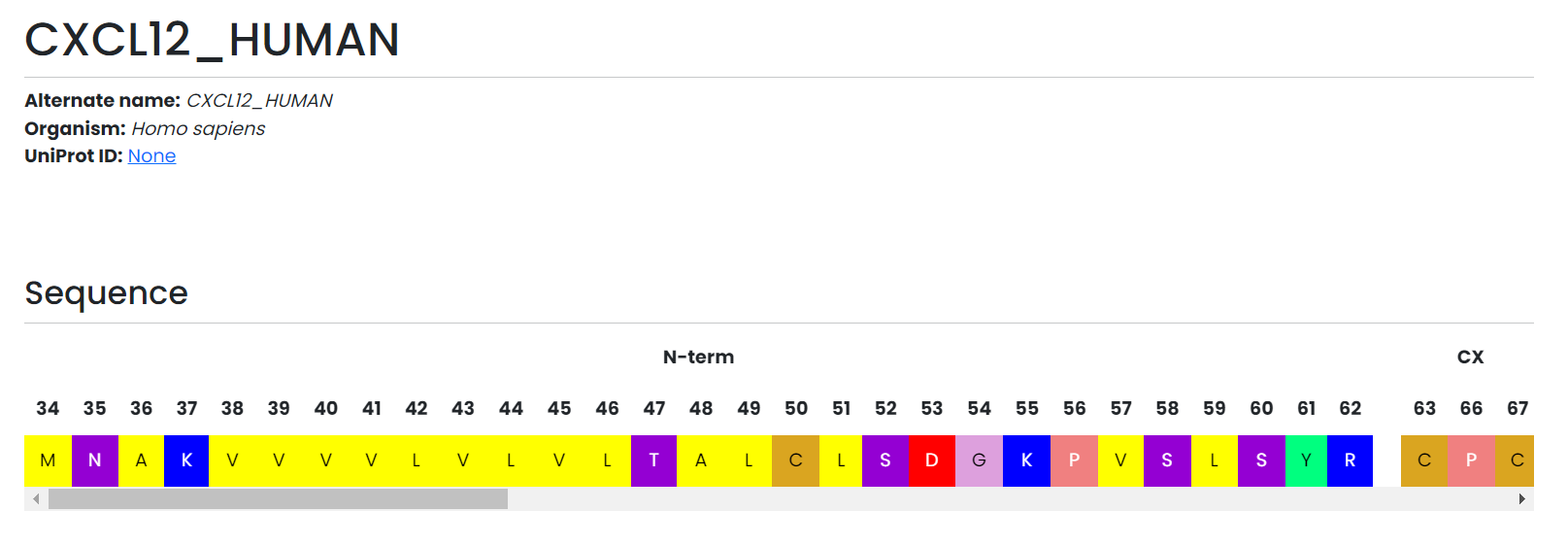
Protein page highlights quick facts, numbering, and interactive diagrams tailored to chemokines.
Structure page
Experimental structure information
- Direct links lead to the PDB entry, UniProt record, and primary publication, keeping trusted references at hand.
- Residue-level alignments compare the experimental chain with the canonical sequence, exportable in CSV or ALN formats.
- Entities and interaction summaries are grouped by chemokine chain so you can spot coverage gaps quickly.
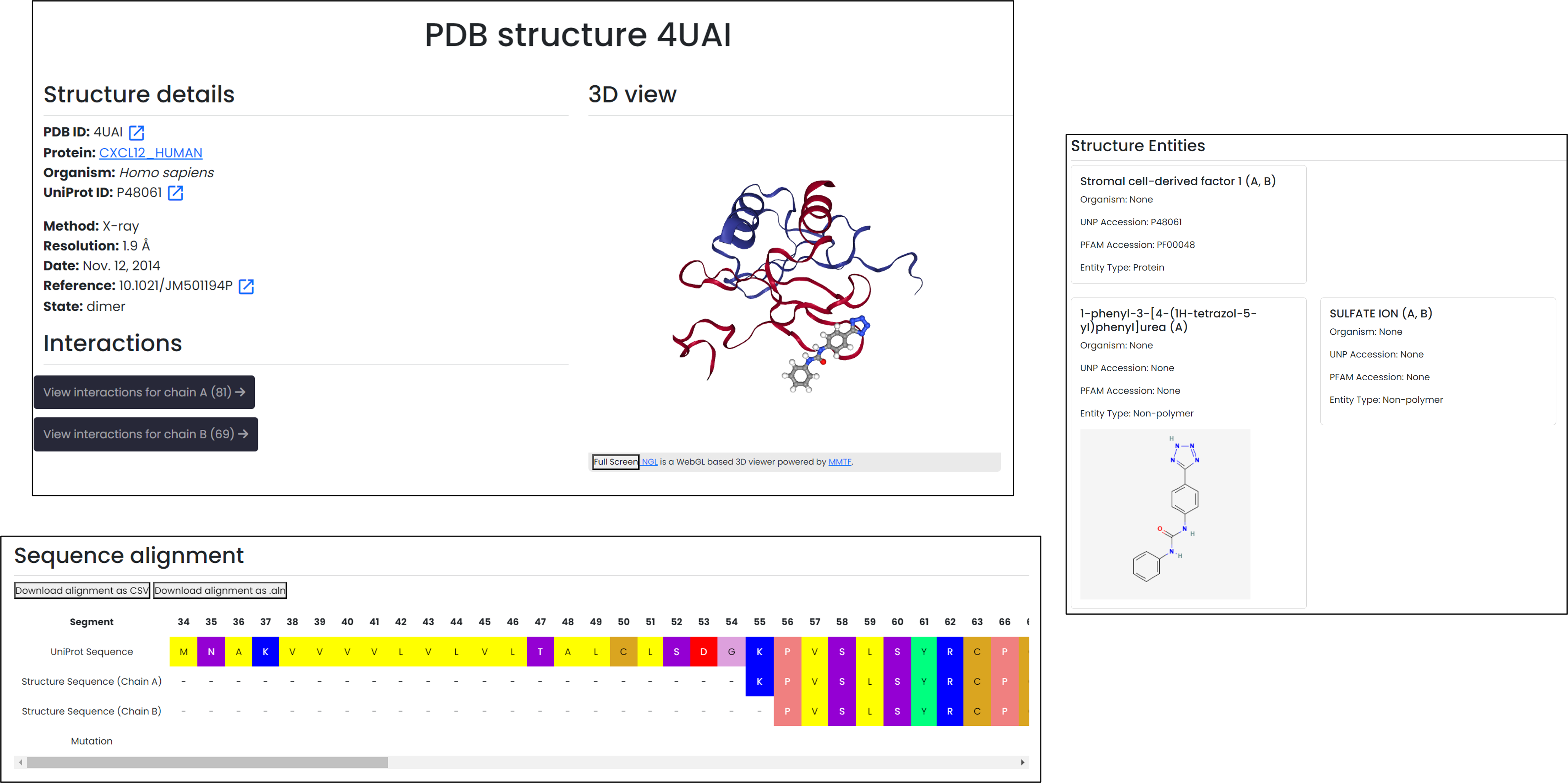
Alignment blocks and entity tables document how each structure supports the CXCL12 entry.
Interaction page
Interactive contacts and fingerprint search
- The embedded 3D viewer renders the chemokine chain in grey and highlights partners so residue contacts are easy to inspect.
- Interaction tables can be exported to Excel, with partner filters to narrow in on specific regions.
- An Interaction Fingerprint (IFP) search computes Tanimoto similarity scores to surface related structures from the ChemoPar-db catalogue.
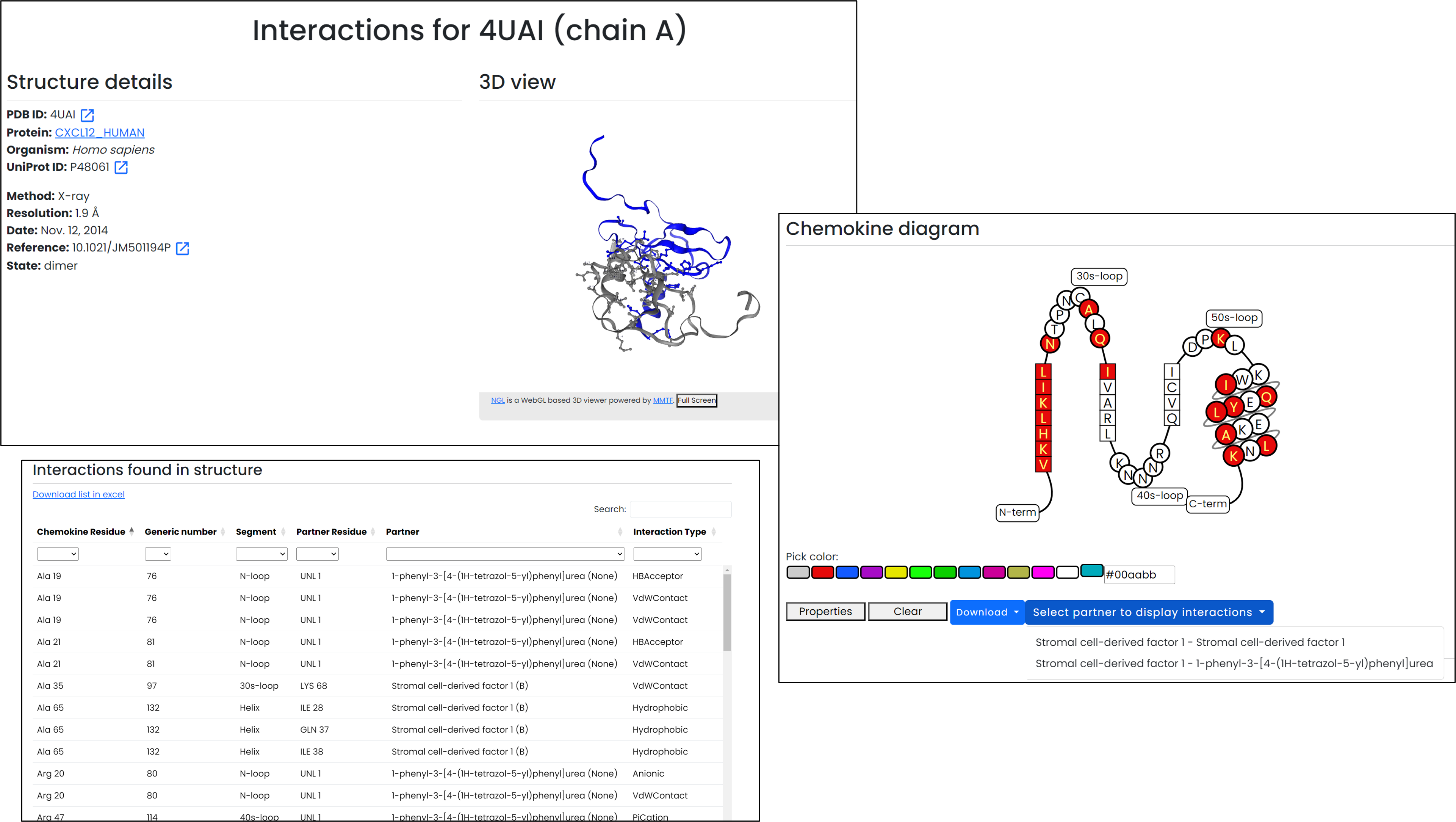
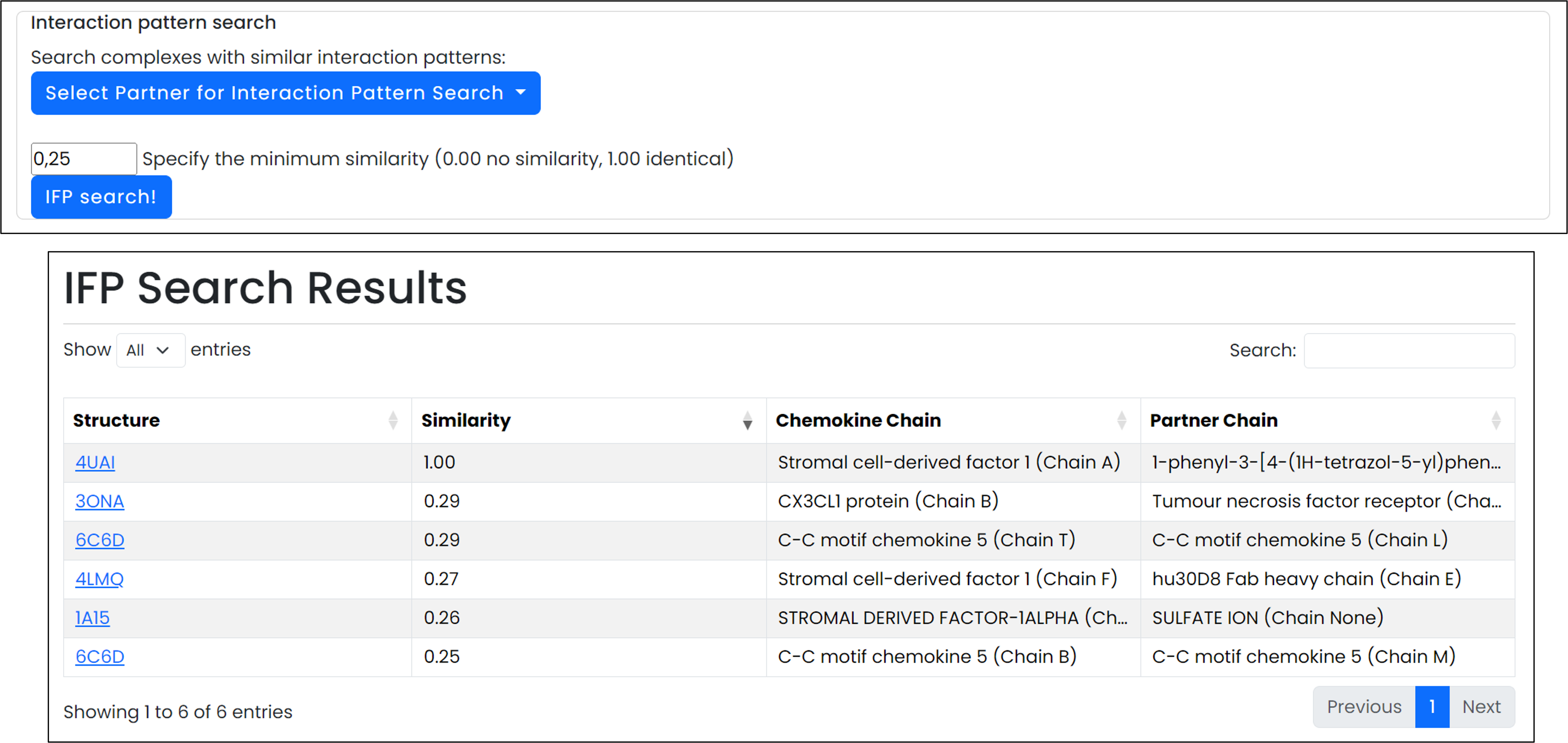
Combine on-page inspection with downloadable fingerprints to compare interaction patterns.
Continue exploring ChemoPar-db
Use the CXCL12 walkthrough as a template for other chemokines — the same numbering, visualisations, and downloads are available site-wide.



